BioXp error-corrected libraries kits are built using proprietary technology that leverages de novo DNA synthesis, error correction, and amplification. The final product is purified library gene fragments ready for downstream applications such as cloning, screening, or selection.
Through the power of push-button, end-to-end automation of the BioXp system, error-corrected libraries are assembled in a single instrument run of less than 16 hours – regardless of design complexities – enabling researchers to reduce synthesis bottlenecks during Design-Build-Test cycles of lead optimization significantly.
Library synthesis time is independent of design complexity.
DNA with the diversity you need & the fidelity you expect
Escape from the design limitations of service providers.
Delivering both fidelity and diversity – from an overnight instrument run
Sequence fidelity increases approximately 2-fold with error correction than without. Median error rate as measured across a panel of representative standard libraries spanning 300-800bp, GC 30-60%.
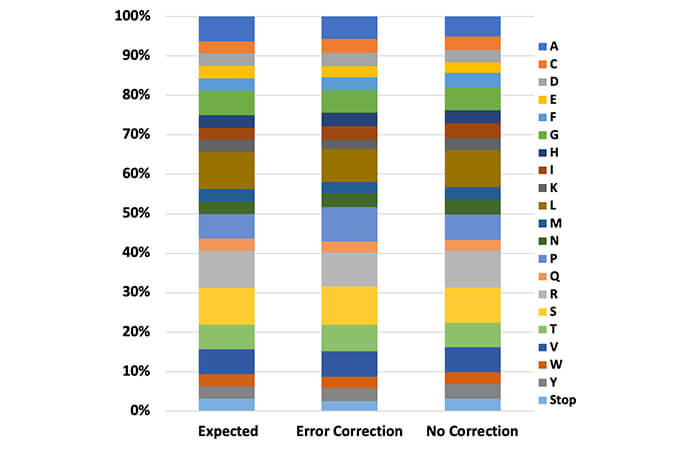
Generation of diversity is unaffected by error-correction technology. Expected vs. observed amino acid distribution analysis from 32 unique NNK degenerate codons with and without error correction.
Median Error Rate: 1 in 5000 bp
BioXp scanning error-corrected libraries offer a wide variety of libraries with varied amino acid sites which can be categorized into simple-scanning (single amino acid/codon substitutions per well) and site-saturation libraries that employ NNN, NNK, and NNS degenerate codons.

* - Lengths of < 2,000 bp may be ordered, but not guaranteed
** - GC content 20% < X < 70% may be ordered, but not guaranteed
BioXp combinatorial error-corrected libraries offer a wide variety of libraries with varied amino acid sites which include NNN, NNK, and NNS degenerate codons as well as degenerate bases from the IUPAC code.

* - Lengths of < 2,000 bp may be ordered, but not guaranteed
** - GC content 20% < X < 70% may be ordered, but not guaranteed